I'll try to cover this paper in three parts:
1. I'll talk about the basic ideas and conclusions in the paper,
2. I'll go over some of the technical details in more depth, so we can see how this group did their analysis,
3. Finally, I'll talk about what I think this does and doesn't imply about evolution - Intelligent Design groups seem ready to jump all over every high-profile paper touching on evolution with one convoluted misinterpretation after another, so we need to deal with potential ways to misread this paper.
So, part one - what this paper is all about:
A group at the Broad Institute (which is part of both MIT and Harvard) has generated about 87 million bases of new gorilla genome sequence; this new sequence data has enabled them to line up large sections of great ape genomes and make an extremely detailed inventory of the DNA base differences among them. They conclude that these results suggest that human and chimp speciation was more complex than previously appreciated, and that there might have been some cross-breeding between the two lineages for some time after they diverged. (And no, this doesn't suggest that humans were having sex with chimps 5 million years ago - there were no humans and chimps then; there were sets of closely related species that might have interbred.)
What I find most exciting about this paper is its application of the power of comparative genomics to primates. Researchers have been comparing the genomes of different yeast species (we have dozens of yeast genomes now), different fly species, etc. and in the process we have learned a lot about both evolution and the basic biology of the cell. Over the last ten years, scientists have developed some powerful computational and statistical tools for doing this kind of analysis; now, we finally have enough primate genome sequence to really start applying these tools to the species we are most interested in - humans!
How do you compare genomes? Even before any genome sequences were available, people were saying that we are "98% chimpanzee." But this figure came from comparing the sequence of a limited number of genes; the catch is that different genes give different answers because genes can evolve at different rates. (In fact, different regions of a gene can evolve at different rate - sections of the gene crucial for a specific function tend to exhibit very few changes, while other regions can evolve fairly rapidly.) Now we can attempt to line up extensive regions of the genome, side by side, and count the number of differences in each region.
Because chimpanzees have their genetic material arranged somewhat differently (for example, some genes which are on chromosome 21 in humans are on chromosome 22 in chimps; you can also have extensive rearrangements within chromosomes), you can't just line up the entire genome from each species and compare them base by base. You have to choose regions that you can line up - regions that haven't undergone extensive rearrangements. In addition, it is helpful to have several species lined up together, to help resolve uncertainties about what kinds of changes took place. Thus, in order to look in detail at human-chimp differences, it is helpful to include genome sequence from gorillas, orangutans, and the much more genetically distant macaques.
In the current paper, the authors were able to line up thousands of regions from chimp, human, gorilla, macaque, and sometimes orangutan genomes, adding up to over thirty million DNA bases that they could directly compare among these species. Previous studies of great apes, according to the paper, covered only about 25 thousand bases.
Once these sequences were lined up, the authors basically counted up the number of bases where the sequences differ (after applying certain filters to eliminate sequence that could confound the analysis - for example, you have to pull out 'hypermutable' regions where the mutation rate is too high to make a valid comparison). The authors could divide the differences into categories - for example, you can have places where:
- the human genome differs from the other four genomes
- the chimp genome differs from the other four genomes
- humans and chimps are the same, but different from the other three
- humans and gorillas are the same, but different from the other three
- chimps and gorillas are the same, but different from the other three
etc...
These researchers found that the divergence (basically how many differences are in a given region) between the human and chimp genomes varies greatly across the genome, but if you take the average for any given chromosome, the divergence for that chromosome is fairly close to the average for the whole genome... but there is a big exception - the X chromosome, which showed a much lower divergence (circled in red in the figure below, from Patterson, et al. - the y-axis is relative divergence, with 1 being the genome average). In other words, human and chimp X-chromosomes are much more similar to each other than expected if the lineages leading to humans and chimps split off from each other 6-7 million years ago.
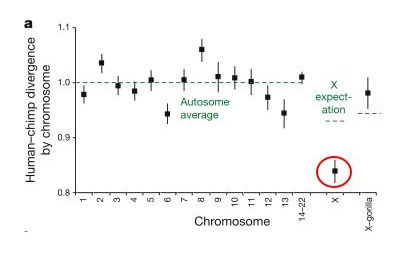
This is the most surprising finding of the study, and it is what leads the authors to suggest that interbreeding occurred between the chimp and human lineages for some time after initially diverging from each other. They suggest that there was an initial split 6-7 million years ago, roughly in line with the fossil record, but that later (less that 6.3 million years ago) there was hybridization between the two lineages resulting in some gene exchange. This paper is still new, so reactions to this scenario are just starting to trickle in, but this hypothesis is the most controversial part of the paper.
It's important to note what is not controversial though: the genomic analysis is fairly standard. The authors have used well-established statistical and computational tools to compare these genomes; the high similarity between human and chimp X chromosomes is real. These genetic analysis techniques are solid, even though some old-school anthropologists and paleontologists still resist them. The challenge now is to decide what this low X divergence is saying about how chimp-human speciation occurred, and that is where the most controversy about this paper will be.
And this is basically what scientists were saying in the NY Times article:
"David Page, a human geneticist at the Whitehead Institute in Cambridge, said the design of the new analysis was "really beautiful, with all the pieces of the puzzle laid out." Whether the hybridization will turn out to be the right solution to the puzzle remains to be seen, "but for the moment I can't think of a better explanation," he said."
In the next post, I'll go into some more technical detail about how these authors did their analysis, as an example of how useful it is to be able to compare multiple genomes.




No comments:
Post a Comment