Molecular Biology: Molecular biologists have long sought to understand the structure and function of the molecular parts of the cell. Like physiologists who study whole organisms, molecular biologists want to know what everything in the cell does. Ideally, we would like to determine the function(s) of every protein made in an organism, the 3-D structure of each of these proteins, the conditions under which these proteins are expressed, how they interact with each other and the non-protein parts of the cell, which portions of our DNA contain regulatory sequences that control gene expression and how those sequences work, etc. etc. In other words, we want to a complete mechanistic picture of the cell.
To even come close to this goal, we must know the DNA sequence of the genome, because every protein and RNA component of the cell is coded by our DNA. By having a genome sequence, we basically have a complete parts list for the cell, even if we don't completely know how to read that list yet. With this parts list, one can make a complete collection of protein-coding genes in an experimentally useful form, and then study those genes systematically. (In a shameless act of self-promotion, I recommend you check out this relevant abstract recently published by our group.) This type of approach has been extensively used in yeast, but can also work for human genes. Another application is the creation of DNA chips, or microarrays, which have been tremendously useful in recent years. Without genome sequences, we could not build these collections and perform such genome-wide experiments.
An advantage of sequencing multiple mammalian genomes (human, chimp, mouse, rat, and now dog), is that we can compare them. This is useful for evolutionary studies (which I'll get to in the next post), but it is also crucial for those purely interested in mechanistic molecular biology. Identifying functionally important portions of a single genome is extremely difficult. Even protein-coding genes are hard to identify. Protein coding genes start with the DNA sequence ATG and end with TAG, TAA, or TGA, with a stretch of DNA in between coding for the amino acids of the protein, like this gene for the human cannabinoid receptor-1, which is the receptor acted on by the major active component of marijuana:
ATGAAGTCGATCCTAGATGGCCTTGCAGATAC
CACCTTCCGCACCATCACCACTGACCTCCTGTACGTGGG
CTCAAATGACATTCAGTACGAAGACATCAAAGGTGACAT
GGCATCCAAATTAGGGTACTTCCCACAGAAATTCCCTTT
AACTTCCTTTAGGGGAAGTCCCTTCCAAGAGAAGATGAC
TGCGGGAGACAACCCCCAGCTAGTCCCAGCAGACCAGGT
GAACATTACAGAATTTTACAACAAGTCTCTCTCGTCCTTC
AAGGAGAATGAGGAGAACATCCAGTGTGGGGAGAACTTC
ATGGACATAGAGTGTTTCATGGTCCTGAACCCCAGCCAG
CAGCTGGCCATTGCAGTCCTGTCCCTCACGCTGGGCACC
TTCACGGTCCTGGAGAACCTCCTGGTGCTGTGCGTCATC
CTCCACTCCCGCAGCCTCCGCTGCAGGCCTTCCTACCAC
TTCATCGGCAGCCTGGCGGTGGCAGACCTCCTGGGGAGT
GTCATTTTTGTCTACAGCTTCATTGACTTCCACGTGTTCC
ACCGCAAAGATAGCCGCAACGTGTTTCTGTTCAAACTGG
GTGGGGTCACGGCCTCCTTCACTGCCTCCGTGGGCAGCC
TGTTCCTCACAGCCATCGACAGGTACATATCCATTCACAG
GCCCCTGGCCTATAAGAGGATTGTCACCAGGCCCAAGGC
CGTGGTGGCGTTTTGCCTGATGTGGACCATAGCCATTGTG
ATCGCCGTGCTGCCTCTCCTGGGCTGGAACTGCGAGAAAC
TGCAATCTGTTTGCTCAGACATTTTCCCACACATTGATGAA
ACCTACCTGATGTTCTGGATCGGGGTCACCAGCGTACTGCT
TCTGTTCATCGTGTATGCGTACATGTATATTCTCTGGAAGG
CTCACAGCCACGCCGTCCGCATGATTCAGCGTGGCACCCAG
AAGAGCATCATCATCCACACGTCTGAGGATGGGAAGGTACA
GGTGACCCGGCCAGACCAAGCCCGCATGGACATTAGGTTAG
CCAAGACCCTGGTCCTGATCCTGGTGGTGTTGATCATCTGCT
GGGGCCCTCTGCTTGCAATCATGGTGTATGATGTCTTTGGGA
AGATGAACAAGCTCATTAAGACGGTGTTTGCATTCTGCAGTA
TGCTCTGCCTGCTGAACTCCACCGTGAACCCCATCATCTATG
CTCTGAGGAGTAAGGACCTGCGACACGCTTTCCGGAGCATGT
TTCCCTCTTGTGAAGGCACTGCGCAGCCTCTGGATAACAGCA
TGGGGGACTCGGACTGCCTGCACAAACACGCAAACAATGCAG
CCAGTGTTCACAGGGCCGCAGAAAGCTGCATCAAGAGCACGG
TCAAGATTGCCAAGGTAACCATGTCTGTGTCCACAGACACGT
CTGCCGAGGCTCTGTGA
The hard part is that not all DNA sequences that fit those criteria are actually protein coding genes - some DNA sequences look like protein coding genes, but they really aren't.
Furthermore, functionally important DNA sequences that do not code for proteins don't start with ATG and end with TAG, TAA or TGA. We can't identify these sites by just looking for certain features in a single genome. This is where genome sequencing is useful - to identify functionally important portions of our genome, whether they code for protein or not, we can compare genomes of related organisms at various evolutionary distances from ours. Because of evolution, genomes change; those parts that are not functionally important can usually change without adverse consequences for the fitness of the organism, while the parts that are functionally important will generally be constrained. So you can compare genomes of various organisms and look for those parts that haven't changed very much; this is a major clue that these parts are authentic functional sequences. If a DNA sequence starts with ATG and ends with TGA, but is poorly conserved among different species, this may not be a true protein coding gene. (On the other hand, it could be a rapidly evolving gene involved in a function like reproduction or the immune system; there are way to test for this.) If a non-coding DNA sequence close to a protein-coding gene is very similar among different species, this sequence could be involved in regulating the nearby protein-coding gene. (Keep in mind, I'm really simplifying things, but this is the basic idea behind comparative genomics.) This is illustrated in the following schematic:
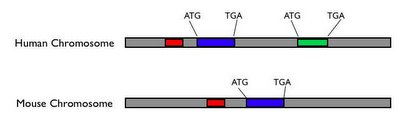
Potential protein-coding genes start with ATG and and with TGA; the blue gene is conserved between human and mouse, while the green gene is not. The blue gene is probably an authentic protein-coding gene, while the green one may not be. The red sequence is not a protein coding sequence, but it is conserved between mouse and human, thus it may be functionally important.
One important problem is choosing the right organisms. Let's say that we're comparing the chimp genome with the human genome and we find a region of DNA that is very similar in the two genomes. It's possible that this region is similar because it is functionally important, or it could be that this region simply hasn't changed much yet because humans and chimps diverged so recently (on an evolutionary scale - 5-6 million years ago). To resolve this issue, we look at the genomes of other organisms, such the mouse or dog. The human and dog genomes diverged much farther back in time, so DNA sequences that are conserved among these two genomes are very likely to be functionally relevant. (On the other hand, the human and dog lineages have diverged enough that some functionally important sequences may not be that similar, which is why we look at an intermediate genome, like mouse or rat.)
This kind of comparison has been very useful in yeast (check out this paper and this one), helping researchers to better define protein-coding genes and non-coding regulatory elements. With this knowledge, we can focus our experimental efforts on systematically characterizing these genomic elements.
In a nutshell, this is one rationale for sequencing genomes. In the next installment, I'll talk about how genomes help us understand evolution, and why the dog genome in particular is useful for studying mammalian evolution.




2 comments:
I know you are on hiatus, but I wanted to say I found this post endlessly fascinating. Hope the move, twins, et al go well for you and your family.
Thanks,
The twins were born Jan. 8; things went wonderfully, I'm thoroughly sleep deprived, and we move next week, but I'll be back.
Post a Comment