Men and women share almost identical genomes - all of us have two copies of chromosomes 1-22, and at least one X chromosome. Differences in genes between men and women are limited to a relatively small number of genes on the Y chromosome. So how do sex differences arise? The answer must be in gender differences in gene regulation. These differences are not only responsible for most of the differences in sex characteristics, but also for many of the gender differences in susceptibility to certain diseases and responses to drugs.
Scientists at UCLA looked at genes in 4 major tissue types that are frequently involved in disease: brain, fat, liver, and muscle tissue. Among the genes that were active in those tissues, they found thousands of genes whose expression was sexually dimorphic, or different in males and females. The most dramatic differences were in liver, fat, and muscle cells, but hundreds of genes also showed expression differences in brain tissue. In brain tissue, 13% of the active genes showed sex differences, while in the other 3 tissue types, more than 50% of active genes showed differences.
Interestingly, sexually dimorphic genes were also genes that tended to be tissue specific. For example, some genes that are active in brain tissue are also active in many other tissues types; other genes are active only in the brain and nowhere else - these latter genes are tissue-specific genes. The researchers found that genes active in brain but nowhere else, or liver but nowhere else, etc., were the genes that showed the greatest differences between males and females. This suggests that most sexually dimorphic genes have highly specialized functions.
Most of the expression differences were small - most genes showed less than a 20% difference in expression level. How significant is this? It's hard to say - in many cases that much of a difference may not matter, but there can be situations where this difference alters the kinetics of a process in the cell and leads to gender differences in drug responses or disease development. Many of the genes that showed differences were enzymes, ion-conducting channels, and cell surface receptors, where small differences in expression could conceivably matter.
What does this all mean? What have we learned from this? Large gene expression studies like this don't always produce groundbreaking, concrete findings. Studies like this are most valuable for the new avenues of research they open up, and the data from this study will provide a resource for future, more specific studies. This is not the first paper to report gender differences in gene expression, but this study does show that these differences are much more extensive than we may have realized before. As I mentioned above, this kind of work lays a foundation for understanding why men and women differ in the incidence and progression of many major diseases.
An important question to ask when you look at the results of any research is, How did the researchers know what they claim to know? As I mentioned, this is not the first study to look at gender specific differences in gene expression, but this study is different in two major ways. The authors used over 300 mice (169 females, 165 males), which increases the statistical power of this study - they are able to reliably detect much smaller differences than previous studies. They also used the offspring of a cross between two fairly different strains of mice, as opposed to earlier studies that worked with one inbred strain. The advantage of this is that they are better able to link specific gene expression differences with certain physiological traits that differ between the two mouse strains. I won't go into the technical reasons why this is true - that's a little too much to deal with in this post.
You may be asking just what exactly are "differences in gene expression" and how do you measure these for thousands of genes at a time? In a study like this one, gene expression is defined as differences in the RNA levels. The level of RNA produced from a DNA-encoded gene is really only a proxy for what actually matters physiologically, the level of protein that is ultimately produced. In many cases, RNA can be a pretty good proxy, and RNA levels are much, much easier to measure because you can measure them on a a microarray. Here is how it works:
Step 1: Kill the mice, get the tissue, grind it up and chemically extract the RNA. Researchers have to be careful at this point to avoid cross contamination - for example, you don't want fat tissue in your muscle sample, although in some cases it's probably impossible to avoid all contamination. Also, in this paper the authors took general tissue types, like whole brain tissue, without separating out sub-types, so their results represent a tissue average and will not capture fine-scale expression differences in specialized parts of the brain.
Step 2: Make fluorescently labeled DNA from the RNA. RNA can act as a template to make DNA, and for reasons that will be clear later, you can use this step to incorporate fluorescent tags. When you use microarrays to look at gene expression, you need two samples for each experiment - a test sample (in this case, tissue from an individual male or female mouse), and a control sample which you can compare the test sample against. In this paper the control consisted of a mixed pool of RNA from both male and female mice, which gives an across-the-board average.
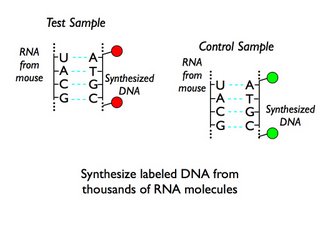
Step 3: Put your fluorescent DNA on the microarray and measure the fluorescence. The fluorescent DNA from the samples matches up, or hybridizes to short segments of DNA that have been printed as spots on a chip; each spot contains DNA for one gene, and a chip containst thousands of genes. The test sample has one kind of fluorescent tag (we'll call it red) and the control sample has another kind (we'll call it green). For each spot you measure how much red fluorescence and how much green fluorescence there is; the readout tells you whether there are more molecules from the test sample or the control sample hybridized to a particular spot. If there is more red on a spot, it means there were more RNA molecules from that gene in the test sample than in the control, and thus that the gene is more highly expressed in the test sample than in the control.
Here is the hybridization at one spot:
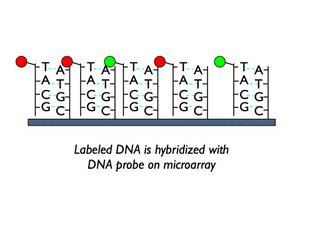
And here is a cartoon version of a microarray (remember, a real one has thousands of spots):

That, in a nutshell, is how you measure gene expression for thousands of genes. Remember, the readout is relative expression - all you can say is how strongly or weakly a gene is expressed in one sample relative to another sample. Measuring absolute levels of RNA requires different technologies, and those technologies haven't yet been adapted for measuring thousands of genes at once.
So after the authors did these experiments for ~300 individual mice (4 samples per mouse - a brain, fat, liver, and muscle sample), they could sit down and analyze their data. One way to represent the data is in a clustering analysis, as shown below:
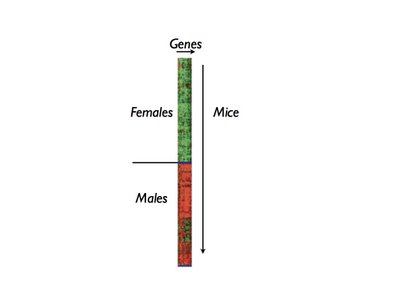
Here is how you read this diagram. It's hard to see this, but the image is made up of several thousand red or green pixels. Each individual pixel represents one individual gene in one individual mouse; the pixel is red if the gene is expressed higher than average in that individual mouse, and green if lower than average. In this plot we are looking at ~40 individual genes, and you can see that these particular genes are expressed higher than average in almost all of the males and lower than average in almost all of the females.
Leaving out a lot of details, that's the general idea of how to measure differences in gene expression on a genome-wide scale.




No comments:
Post a Comment